Lorentz force
We simulate the motion of a charged particle exposed to an electric and magnetic field. The situation corresponds to that encountered in a vacuum tube with a long filament surrounded by a cylindrical anode with a solenoid placed on top. This is more complicated than the usual discussion of a charged particle with constant velocity (acquired, e.g., upon acceleration in an electric field) entering a homogeneous magnetic field (circular trajectory) or an electric field aligned with the magnetic field (helical motion as a result of independent influence of the fields on the position variables). In this example the motion is confined to a plane, the particles are accelerated by an electric field acting in the radial direction. Once they acquire a non-zero velocity the Lorentz force deflects the particle away from the radial path.
The Verlet algorithm used for the Coulomb problem requires a modification to account for the velocity dependence of the Lorentz force. The equations of motion are written in Cartesian coordinates (E and B are the magnitudes of the electric field acting in the radial direction and the magnetic field acting in the z direction respectively). The radial dependence of the electric field strength was determined from a solution to the Laplace equation in cylindrical coordinates (see Laplace.mws ), or from Gauss' law ( Gauss.mws ).
m diff(x(t),t$2) = q [E*x(t)/(x(t)^2+y(t)^2) + vy(t)*B]
m diff(y(t),t$2) = q [E*y(t)/(x(t)^2+y(t)^2) - vx(t)*B]
> with(linalg):
Warning, the protected names norm and trace have been redefined and unprotected
The Cartesian components for the Lorentz force are calculated as follows:
> v:=vector([vx,vy,vz]);
![]()
> Bf:=vector([Bx,By,Bz]);
![]()
> crossprod(v,Bf);
![]()
Now we restrict the homogeneous magnetic field to be aligned with the z axis:
> subs(Bx=0,By=0,%);
![]()
We choose the mass (units) and charge sign:
> m:=1; q:=-1;
![]()
![]()
We worry about the step-size for small distances r as the force is inversely proportional to r. We should try various small values.
> dt:=0.05; dt2:=dt^2;
![]()
![]()
> vx0:=0; vy0:=0;
![]()
![]()
We have to start the trajectories at a finite distance (filament thickness or larger). We can also choose finite initial velocities (they depend on the temperature of the filament).
Let's measure distances in multiples of the filament radius.
> rx[0]:=1; rx[1]:=rx[0]+dt*vx0;
![]()
![]()
> ry[0]:=1; ry[1]:=ry[0]+dt*vy0;
![]()
![]()
> E:='E'; B:='B';
![]()
![]()
The electric field varies with distance according to the following functions. To obtain the electric force one needs to multiply with the charge of the probe particle (electron).
> Ex:=-x/(x^2+y^2);
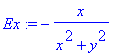
> Ey:=-y/(x^2+y^2);
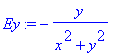
> Verlet:=proc(n1,E0,B0) local Fx,Fy,vx,vy; global rx,ry,dt,dt2,m,q,Ex,Ey;
> vx:=(rx[n1]-rx[n1-1])/dt; vy:=(ry[n1]-ry[n1-1])/dt;
> Fx:=evalf(q*subs(x=rx[n1],y=ry[n1],E0*Ex)+q*B0*vy);
> Fy:=evalf(q*subs(x=rx[n1],y=ry[n1],E0*Ey)-q*B0*vx);
> rx[n1+1]:=2*rx[n1]-rx[n1-1]+dt2/m*Fx;
> ry[n1+1]:=2*ry[n1]-ry[n1-1]+dt2/m*Fy;
> end:
> N:=200;
![]()
> for i from 1 to N do:
> Verlet(i,1,2); od:
> nops(convert(rx,list));
![]()
> rx[40],ry[40];
![]()
> plot([seq([rx[i],ry[i]],i=0..N)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce22.gif)
This doesn't seem right. Reducing the time step shows a repetitive motion.
How can we fix the equations to use the velocities implicitly? Let us write the recursions:
> rx:='rx': ry:='ry': dt:='dt': q:='q': i:='i': m:=1: B:='B': f:='f': g:='g': rx_ip1:='rx_ip1': ry_ip1:='ry_ip1':
> eq1:=m*(rx_ip1+rx[i-1]-2*rx[i])/dt^2=f+B*(ry_ip1-ry[i-1])/(2*dt);
![eq1 := (rx_ip1+rx[i-1]-2*rx[i])/(dt^2) = f+1/2*B*(r...](images/LorentzForce23.gif)
> eq2:=m*(ry_ip1+ry[i-1]-2*ry[i])/dt^2=g-B*(rx_ip1-rx[i-1])/(2*dt);
![eq2 := (ry_ip1+ry[i-1]-2*ry[i])/(dt^2) = g-1/2*B*(r...](images/LorentzForce24.gif)
> sol:=solve({eq1,eq2},{rx_ip1,ry_ip1});
![sol := {rx_ip1 = (-4*rx[i-1]+rx[i-1]*B^2*dt^2+8*rx[...](images/LorentzForce25.gif)
![sol := {rx_ip1 = (-4*rx[i-1]+rx[i-1]*B^2*dt^2+8*rx[...](images/LorentzForce26.gif)
> assign(sol);
> rx_ip1;
![(-4*rx[i-1]+rx[i-1]*B^2*dt^2+8*rx[i]+4*f*dt^2-4*B*d...](images/LorentzForce27.gif)
Now redefine Verlet:
> Verlet:=proc(n1,E0,B0) local Fx,Fy; global rx,ry,dt,dt2,m,q,Ex,Ey,rx_ip1,ry_ip1;
> Fx:=evalf(q*subs(x=rx[n1],y=ry[n1],E0*Ex));
> Fy:=evalf(q*subs(x=rx[n1],y=ry[n1],E0*Ey));
> rx[n1+1]:=subs(f=Fx,g=Fy,B=B0,rx_ip1);
> ry[n1+1]:=subs(f=Fy,g=Fy,B=B0,ry_ip1);
> end:
> q:=-1;
![]()
> dt:=0.05; dt2:=dt^2;
![]()
![]()
> vx0:=0; vy0:=0;
![]()
![]()
> rx[0]:=1; rx[1]:=rx[0]+dt*vx0;
![]()
![]()
> ry[0]:=1; ry[1]:=ry[0]+dt*vy0;
![]()
![]()
Now we carry out a large number of steps.
> N:=1000;
![]()
> for i from 1 to N do:
> Verlet(i,1,2); od:
> nops(convert(rx,list));
![]()
> rx[4],ry[4];
![]()
> plot([seq([rx[i],ry[i]],i=0..N)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce40.gif)
We see that the motion is that of a regularly shaped rosette. To achieve better accuracy (we note that the solution is slipping after a complete round) we would need to reduce the step-size.
We can try cases where the solution gets further away from the filament:
> dt:=0.02;
![]()
> N:=5000;
![]()
> for i from 1 to N do:
> Verlet(i,10,1/2); od:
> plot([seq([rx[i*10],ry[i*10]],i=1..N/10)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce43.gif)
Is this solution accurate? Let us re-compute with half the step-size:
> dt:=0.01;
![]()
> N:=10000;
![]()
> for i from 1 to N do:
> Verlet(i,10,1/2); od:
> plot([seq([rx[i*20],ry[i*20]],i=1..N/20)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce46.gif)
Now we should be concerned:
> dt:=0.005;
![]()
> N:=20000;
![]()
> for i from 1 to N do:
> Verlet(i,10,1/2); od:
> plot([seq([rx[i*40],ry[i*40]],i=1..N/40)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce49.gif)
It appears as if the motion should be quite regular, i.e., the 'circles' should be progressing steadily in the x-y plane.
> dt:=0.002;
![]()
> N:=50000;
![]()
> for i from 1 to N do:
> Verlet(i,10,1/2); od:
> plot([seq([rx[i*100],ry[i*100]],i=1..N/100)],style=point,scaling=constrained);
![[Maple Plot]](images/LorentzForce52.gif)
We seem to be getting a different answer each time...
> dt:=0.001;
> N:=100000;
> for i from 1 to N do:
> Verlet(i,10,1/2); od:
> plot([seq([rx[i*200],ry[i*200]],i=1..N/200)],style=point,scaling=constrained);
![]()
![]()
![[Maple Plot]](images/LorentzForce55.gif)
Now that we have a satisfying answer (which takes a long time to compute) we turn to Maple's dsolve[numeric] engine. It can handle the solution much more quickly, as it does not use a fixed step-size. It slows down in the time-step only when the charged particle returns to the radial distance r =1, where we expect intuitively the solution to be most difficult to achieve.
>
> restart;
> q:=-1; m:=1: E0:=1; B0:=2;
![]()
![]()
![]()
> de1:=m*diff(x(t),t$2)=-q*E0*x(t)/(x(t)^2+y(t)^2)+diff(y(t),t)*q*B0;
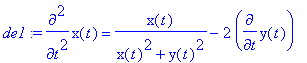
> de2:=m*diff(y(t),t$2)=-q*E0*y(t)/(x(t)^2+y(t)^2)-diff(x(t),t)*q*B0;
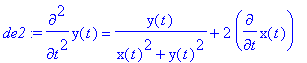
> #dsolve({de1,de2,x(0)=1,D(x)(0)=0,y(0)=1,D(y)(0)=0},{x(t),y(t)});
Maple tries for a long time to obtain an analytic solution - to no avail. The pair of ODEs is nonlinear.
> sol:=dsolve({de1,de2,x(0)=1,D(x)(0)=0,y(0)=1,D(y)(0)=0},{x(t),y(t)},numeric,output=listprocedure):
> X:=subs(sol,x(t)): Y:=subs(sol,y(t)):
> plot(['X(t)','Y(t)',t=0..40],scaling=constrained,thickness=2);
![[Maple Plot]](images/LorentzForce61.gif)
It pays off to use a sophisticated differential equation solver. Let us try the other case:
> restart;
> q:=-1; m:=1: E0:=10; B0:=1/2;
![]()
![]()
![]()
> de1:=m*diff(x(t),t$2)=-q*E0*x(t)/(x(t)^2+y(t)^2)+diff(y(t),t)*q*B0;
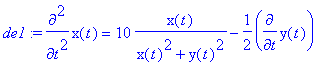
> de2:=m*diff(y(t),t$2)=-q*E0*y(t)/(x(t)^2+y(t)^2)-diff(x(t),t)*q*B0;
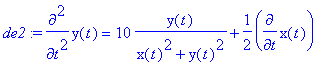
> sol:=dsolve({de1,de2,x(0)=1,D(x)(0)=0,y(0)=1,D(y)(0)=0},{x(t),y(t)},numeric,output=listprocedure):
> X:=subs(sol,x(t)): Y:=subs(sol,y(t)):
> plot(['X(t)','Y(t)',t=0..200],scaling=constrained,thickness=2);
![[Maple Plot]](images/LorentzForce67.gif)
For very long integration times Maple's built-in numerical method may break down as well. One can verify the accuracy by trying different numerical methods, such as lsode or dverk78 instead of the usual adaptive Runge-Kutta-Fehlberg 4/5 method (cf the help pages on dsolve[numeric] ).
> sol:=dsolve({de1,de2,x(0)=1,D(x)(0)=0,y(0)=1,D(y)(0)=0},{x(t),y(t)},numeric,method=dverk78,output=listprocedure):
> X:=subs(sol,x(t)): Y:=subs(sol,y(t)):
> plot(['X(t)','Y(t)',t=0..200],scaling=constrained,thickness=2);
![[Maple Plot]](images/LorentzForce68.gif)
>