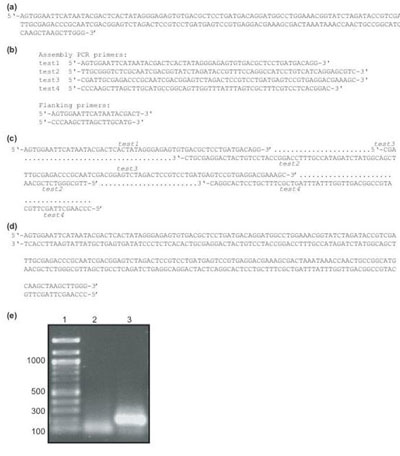
Protocol for assembly PCR reactionThe program was experimentally verified by using the oligodeoxynucleotides determined by the program for the two-step assembly PCR construction of a DNA molecule that is to be used to produce an RNA molecule. The desired RNA product is a 191-nucleotide moleculeconsisting of 5’ and 3’ cis hammerhead ribzoymes and a core 20 nucleotide region that forms a hairpin structure and is having its structure studied in our lab by nuclear magnetic resonance (NMR) methods. The program broke this 191-nucleotide DNA molecule into four segments for the first PCR reaction and produced the two oligodeoxynucleotide molecules for the second PCR reaction (Figure 2.a, b, c, d). Upon receipt, the oligodeoxynucleotides for the first step of assembly PCR were diluted to 0.125 µg/µL with double distilled water, while the oligodeoxynucleotides for the second PCR step were diluted to 0.25 µg/µL. For the first PCR reaction, 4 µL of each oligo, 4 µL of 5 mM dNTPs, 10 µL of 10x thermopol buffer (NEB), 1.5 µL of Vent DNA polymerase (2000 U/mL), and 68.5 µL of double distilled water were combined. This mixture was then subjected to 8 cycles of amplification at 94 °C (1.5 min), 54 °C (2 min), and 72 °C (3 min). During the first cycle, the 94 °C step was performed for 7 min. After the last cycle completed, an additional 5 min 72 °C elongation step was performed. For the second PCR reaction, 1 µL of the crude mixture from the first PCR reaction was mixed with 4 µL of each primer, 4 µL of 5 mM dNTPs, 10 µL of 10x thermopol buffer (NEB), 1.5 µL of Vent DNA polymerase, and 75.5 µL of double distilled water. This mixture was then subjected to 25 cycles of amplification. Each cycle consisted of a 30 second 94°C step, a 2 min 54 °C step, and a 1.5 min 72 °C step. Prior to the first cycle, a 5 min 94 °C step was used. A 5 min 72 °C elongation step was included following the final cycle. The PCR mixtures were analyzed by agarose gel electrophoresis. For each reaction a 6 µL sample was mixed with 2 µL of blue-green dye. The gel was stained with ethidium bromide for 20 minutes, and observed under UV light. As shown by gel analyses (Figure 2.e), the first PCR reaction produces a diffuse band or smear, while the desired full length product results from the second PCR reaction. This behavior is consistent with previous reports of assembly PCR gene construction. The product of the second PCR reaction was cloned in to the pUC18 plasmid, and the correctness of its sequence was verified by DNA sequencing.
Figure 2. The construction of a 191-nucleotide DNA molecule using the oligodeoxynucleotides determined by the Assembly PCR Oligo Maker program. (a) The sequence of the 191-nucleotide DNA target to be produced. (b) The DNA sequences reported by Assembly PCR Oligo Maker program. Sequences for both step of the assembly PCR reaction are reported. (c) Diagram showing how the four oligodeoxynucleotides anneal to produce the full-length dsDNA product (d) The final dsDNA product produced by the assembly PCR reaction (e) Agarose gel showing the results of the first (lane 2) and second (lane 3) PCR steps. The desired 191-nucleotide molecule is visible after the second step PCR step. DNA length markers are shown in lane 1. |