� Fractal Dimension �
��
�� The analysis of Poincare plots for the Duffing oscillator which is explored in � Duffing.mws � indicates that for certain parameter ranges the Poincare section represents an accumulation of points. Our aim is to calculate the fractal dimension of this strange attractor. We introduce a simple box counting algorithm in order to calculate the capacity dimension, and we test it on a fractal structure whose dimension can be derived from the algorithm that prescribes it. First, we have to construct an algorithm that generates this fractal, which is called a Koch curve. �
��
�� Koch curve �
��
�� A Koch curve can be generated by an iterative algorithm. A basic step consists of taking a unit line, cutting it in three, and replacing the middle segment by an equilateral triangle without base, thereby extending the length of the original entire line segment by 4/3. �
��
�� Suppose we define the curve by its vertices. We implement the algorithm by the following steps: �
�� 1) find the points that cut the line into three segments; �
�� 2) find the midpoint, and rotate it to horizontal orientation, displace the y-coordinate by the required amount; �
�� 3) rotate the shifted midpoint back to the original orientation; �
�� 4) assemble a list that contains all required vertices. �
��
�� This may not be the most efficient implementation, but it is starightforward to understand. �
��
��
| > | �restart; with(stats): # statistics package is used for least-squares fit. � |
�
| > | �Koch:=proc(LL) local xi,yi,xf,yf,x1,y1,x2,y2,xh,yh,phi,cf,sf,x3,y3,xhp,yhp,d; � |
�
| > | �xi:=LL[1][1]: yi:=LL[1][2]: xf:=LL[2][1]: yf:=LL[2][2]: � |
�
| > | �x1:=evalf(xi+(xf-xi)/3): y1:=evalf(yi+(yf-yi)/3): � |
�
| > | �x2:=evalf(xi+2*(xf-xi)/3): y2:=evalf(yi+2*(yf-yi)/3): � |
�
| > | �xh:=evalf((xf+xi)/2): yh:=evalf((yf+yi)/2): � |
�
| > | �phi:=evalf(arctan(yf-yi,xf-xi)); � |
�
| > | �cf:=cos(phi): sf:=sin(phi): � |
�
| > | �xhp:=xh*cf+yh*sf: yhp:=-xh*sf+yh*cf: d:=evalf(sqrt((xf-xi)^2+(yf-yi)^2)): � |
�
| > | �yhp:=evalf(yhp+d/6*sqrt(3)): � |
�
| > | �x3:=evalf(xhp*cf-yhp*sf): y3:=evalf(xhp*sf+yhp*cf): � |
�
| > | �[LL[1],[x1,y1],[x3,y3],[x2,y2],LL[2]]: end: � |
�
| > | �Koch([[0,0],[0,1]]); � |
�
![]() �
�
�
| > | �Koch([[0,0],[1,0]]); � |
�
![]() �
�
�
| > | �Koch([[0,0],[1,1]]); � |
�
![]() �
�
�
| > | �plot(%,scaling=constrained,axes=boxed); � |
�
![[Maple Plot]](images/FractalDimension4.gif) �
�
� Now we need a driver: we want to take a list of points, and for each pair of subsequent points we want to apply the Koch procedure. �
��
| > | �DriveK:=(LL)-> � |
�
| > | �[seq( op(Koch([LL[i],LL[i+1]])),i=1..nops(LL)-1 )]: � |
�
| > | �DriveK([[0,0],[1,0]]); � |
�
![]() �
�
�
| > | �DriveK(%); � |
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
�
| > | �plot(%,scaling=constrained,axes=boxed); � |
�
![[Maple Plot]](images/FractalDimension10.gif) �
�
� The Koch curve is an important exercise in order to understand fractal dimension, because the increase in length of the figure after each step is obvious: a factor of 4/3 applies for each iteration. Let us verify this numerically: �
��
| > | �Length:=LL->evalf(add(sqrt((LL[i+1][1]-LL[i][1])^2+(LL[i+1][2]-LL[i][2])^2),i=1..nops(LL)-1)); � |
�
![Length := proc (LL) options operator, arrow; evalf(add(sqrt((LL[i+1][1]-LL[i][1])^2+(LL[i+1][2]-LL[i][2])^2),i = 1 .. nops(LL)-1)) end proc](images/FractalDimension11.gif) �
�
�
| > | �Length(Koch([[0,0],[0,1]])); � |
�
![]() �
�
�
| > | �Length((DriveK@@2)([[0,0],[0,1]])); � |
�
![]() �
�
�
| > | �evalf((4/3)^2); � |
�
![]() �
�
�
| > | �plot((DriveK@@5)([[0,0],[1,0]]),scaling=constrained,axes=boxed); � |
�
![[Maple Plot]](images/FractalDimension15.gif) �
�
� The capacity dimension of the Koch curve is given as: �
��
| > | �D_Koch:=evalf(log(4)/log(3)); � |
�
![]() �
�
� Why is this so? Do some reading on fractal dimension. �
��
�� How rapidly does the number of points grow with each layer? �
��
| > | �seq(nops((DriveK@@i)([[0,0],[1,0]])),i=1..5); � |
�
![]() �
�
� In order to implement the box counting method we need to pick a pixel size resolution and connect the points. In a plotting program this is carried out by the graphics engine (in the � plot � command above the default method is to connect the points by a straight line). Redraw the above graph using � style=point,symbol=point � to observe what the plot engine has done for us. Of course, we can simply iterate the curve to a sufficiently high degree such that at a given smallest box size resolution (e.g., pixel size for a graphics format) all points are connected. �
��
��
| > | �K7:=(DriveK@@7)([[0,0],[1,0]]): � |
�
| > | �K8:=DriveK(K7): � |
� The two data sets K7 and K8 allow us to experiment below. Let us choose the smaller one: �
��
| > | �K_u:=K7: � |
�
| > | �k_max:=nops(K_u); � |
�
![]() �
�
� Choose a maximum value for the resolution layer (smallest box size) at which the plane with the curve enclosed will be explored. The boxes are squares in our case, because the points that make up the Koch curve are two-dimensional. The total area in which the fractal resides is a rectangle: we can account for that. We are setting up an algorithm analogous to a histogram calculation except that we don't care how many times a given bin has been occupied, we just determine whether it has been hit, and then add up how many hits we get at a certain level of resolution. The baseline for a bin (square) is called � epsilon=1/n � , where n is the resolution layer, and we need to look at the log of the total number of box hits versus � log(1/epsilon) � ; the slope of this graph gives the fractal dimension (in the limit of � epsilon � going to zero). At the lowest layer of resolution (n=1) we use 10 times 5 boxes. �
��
| > | �layer:=15; � |
�
![]() �
�
�
| > | �LXY:=[]: � |
�
| > | �for n from 1 to layer do: � |
�
| > | �for i from 1 to (10*n) do: � |
�
| > | �for j from 1 to (5*n) do: � |
�
| > | �P[i,j]:=0; � |
�
| > | �od: � |
�
| > | �od: � |
�
| > | �Pepsilon:=1/n: � |
�
| > | �Pboxcount:=0: � |
�
| > | �for k from 1 to k_max do: ix:=trunc(10*n*(K_u[k][1]+0.5)); iy:=trunc(5*n*(K_u[k][2]+0.25)); � |
�
| > | �if (P[ix,iy]=0) then � |
�
| > | �P[ix,iy]:=1: � |
�
| > | �Pboxcount:=Pboxcount + 1: � |
�
| > | �fi: � |
�
| > | �od: print("For res. ",n,"find ",Pboxcount," hits out of ",5*n*10*n," boxes"); � |
�
| > | �LXY:=[op(LXY), [evalf(log10(1/Pepsilon)),evalf(log10(Pboxcount))]]: � |
�
| > | �od: � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �PL1:=plot(LXY,style=point): plots[display](PL1); � |
�
![[Maple Plot]](images/FractalDimension35.gif) �
�
� When the number of boxes is small we have no useful results, so let us ignore them in the regression analysis: �
��
| > | �nL:=4; nU:=nops(LXY); � |
�
![]() �
�
�
![]() �
�
�
| > | �sol:=fit[leastsquare[[x,y]]]([[seq(LXY[i][1],i=nL..nU)],[seq(LXY[i][2],i=nL..nU)]]); � |
�
![]() �
�
�
| > | �FractDim:=coeff(rhs(sol),x); � |
�
![]() �
�
�
| > | �PL2:=plot(rhs(sol),x=0.5..1.2,color=blue): plots[display](PL1,PL2); � |
�
![[Maple Plot]](images/FractalDimension40.gif) �
�
� Exercise 1: �
�� Push the calculation to higher levels of resolution. What do you find? Then use the larger data sample ( � K8 � ) and repeat the analysis. For a better analysis you want to use a regression fit with uncertainty estimate of the coefficients. An algorithm for this is provided in the worksheet � DataAnalysis.mws � . �
��
�� Duffing Attractor �
��
��
�� The Duffing oscillator has been introduced in � Duffing.mws � and it has been shown that for certain parameter values the limit cycle becomes an interesting structure. Here we carry out a measurement of a capacity dimension to demonstrate the fractal nature of the attractor, which is therefore called a strange attractor. In maple8 the integration in dsolve[numeric] has become fast enough that we can extract useful information without having to resort to C-coded routines (such as the method=dna option, which requires a special library to be loaded). For maple7 we still recommend the usage of the DNA package; for maple8 it was unavailable yet at the beginning of September, 2002). �
��
�� For maple8 (without DNA) we restrict the precision to 13 digits. Going beyond 14 digits results in a massive slow-down. �
��
| > | �restart; Digits:=13; � |
�
![]() �
�
�
| > | �unprotect(gamma): with(plots): with(stats): � |
�
Warning, the name changecoords has been redefined
�
�
�
| > | �gamma:=1/10: eta:=1: delta:=1/4: � |
�
| > | �plot(-1/2*eta*x^2+delta*x^4/4,x=-3..3,title="Potential Energy"); � |
�
![[Maple Plot]](images/FractalDimension42.gif) �
�
� We include a periodic driving force (amplitude � F0 � and circular frequency � nu � ) and start the oscillator from the unstable equilibrium point: �
��
| > | �nu:=2: F0:=2.5: DF:=F0*sin(nu*t); � |
�
![]() �
�
�
| > | �DE:=diff(x(t),t)=y(t),diff(y(t),t)=-gamma*y(t)+eta*x(t)-delta*x(t)^3+DF; � |
�
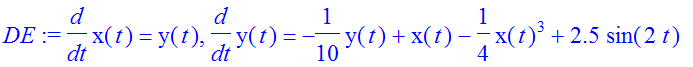 �
�
�
| > | �IC:=x(0)=0,y(0)=0; #start at the unstable equilibrium point � |
�
![]() �
�
� In case you haven't looked at Duffing.mws: here is a sample trajectory: �
��
| > | �#sol:=dsolve({DE,IC},{x(t),y(t)},numeric,output=listprocedure): � |
�
| > | �#X:=subs(sol,x(t)): Y:=subs(sol,y(t)): � |
�
| > | �#plot('X(t)',t=0..100,numpoints=2000,title="Trajectory"); � |
�
| > | �#plot(['X(t)','Y(t)',t=0..50],numpoints=1000,title="Phase-space Diagram"); � |
� The Poincare section is obtained by a stroboscopic view at intervals spaced apart by the period of the driving force. An interesting question arises: what happens to the Poincare section when we change the phase at which we record the phase-space point? It turns out that one gets a similar structure. It is possible to record these different sequences, and run an animation. We do this before we proceed to compute the fractal dimension, just to convince ourselves that there is no loss of generality by fixing the phase at which we record the section. �
��
| > | �tau:=evalf(2*Pi/nu); LL:='LL': � |
�
![]() �
�
�
| > | �t:='t': sol:=dsolve({DE,IC},{x(t),y(t)},numeric,output=listprocedure): � |
�
| > | �X:=subs(sol,x(t)): Y:=subs(sol,y(t)): � |
�
| > | �dtau:=tau/10; � |
�
![]() �
�
�
| > | �for i from 1 to 2000 do: for j from 1 to 10 do: t_i:=evalf(i*tau+(j-1)*dtau); LL||j[i]:=[X(t_i),Y(t_i)]: od: od: � |
�
| > | �for j from 1 to 10 do: � |
�
| > | �PL[j]:=plot([seq(LL||j[i],i=1..2000)],style=point,symbol=point,axes=boxed,view=[-5..5,-8..8],title="Poincare Section",color=blue): od: � |
�
| > | �display([seq(PL[j],j=1..10)],insequence=true); � |
�
![[Maple Plot]](images/FractalDimension48.gif) �
�
� Exercise 2: �
�� Explore the behaviour of this Duffing dust for small variations in the drive amplitude and frequency. What is your guess for the fractal dimension? �
��
�� Now we collect our data sample for fractal dimensions analysis. We record in excess of 100,000 points. This takes some time (under 10 minutes on a > 1GHz machine) �
��
| > | �t:='t': sol:=dsolve({DE,IC},{x(t),y(t)},numeric,output=listprocedure): � |
�
| > | �X:=subs(sol,x(t)): Y:=subs(sol,y(t)): � |
�
| > | �for i from 1 to 200000 do: t_i:=i*tau; LL[i]:=[X(t_i),Y(t_i)]: od: � |
�
�� To analyze the section we adapt the box counting method. We should perform the analysis with varying level of layers and increasing point number � k_max � (the maximum possible value being limited to the pre-computed maximum). We start with 200,000 points, and can repeat the exploration with larger samples (requires a re-run of the previous loop for longer times). �
��
�� The adjustment to the box counting method concerns a shift in the x- and y-coordinates by 5 and 8 units respectively, and a choice of resolution (rectangle). �
��
| > | �k_max:=200000; � |
�
![]() �
�
�
| > | �layer:=20; � |
�
![]() �
�
�
| > | �LXY:=[]: � |
�
| > | �for n from 1 to layer do: � |
�
| > | �for i from 1 to (10*n) do: � |
�
| > | �for j from 1 to (12*n) do: � |
�
| > | �P[i,j]:=0; � |
�
| > | �od: � |
�
| > | �od: � |
�
| > | �Pepsilon:=1/n: � |
�
| > | �Pboxcount:=0: � |
�
| > | �for k from 1 to k_max do: ix:=trunc(n*(LL[k][1]+5)); iy:=trunc(n*(LL[k][2]+8)); � |
�
| > | �if (P[ix,iy]=0) then � |
�
| > | �P[ix,iy]:=1: � |
�
| > | �Pboxcount:=Pboxcount + 1: � |
�
| > | �fi: � |
�
| > | �od: print("For res. ",n,"find ",Pboxcount," hits out of ",12*n*10*n," boxes"); � |
�
| > | �LXY:=[op(LXY), [evalf(log10(1/Pepsilon)),evalf(log10(Pboxcount))]]: � |
�
| > | �od: � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �PL1:=plot(LXY,style=point): display(PL1); � |
�
![[Maple Plot]](images/FractalDimension71.gif) �
�
� The numbers and the graph tell us two things (at least): �
��
�� 1) early on for relatively large boxes the box count increases with a slope of approximately 1.6; �
�� 2) later the slope settles to about 0.6, and epsilon is beginning to be small, i.e., we should trust those results. �
�� 3) clearly, for the last few points we are hitting the limit: the number of boxes becomes comparable to the number of points. �
��
�� So let us extract a slope: �
��
| > | �nL:=10; nU:=nops(LXY)-2; � |
�
![]() �
�
�
![]() �
�
�
| > | �sol:=fit[leastsquare[[x,y]]]([[seq(LXY[i][1],i=nL..nU)],[seq(LXY[i][2],i=nL..nU)]]); � |
�
![]() �
�
�
| > | �PL2:=plot(rhs(sol),x=0..1.5,color=blue): display(PL1,PL2); � |
�
![[Maple Plot]](images/FractalDimension75.gif) �
�
�
| > | �FractDim:=coeff(rhs(sol),x); � |
�
![]() �
�
� So it appears as if the attractor is indeed made of dust, i.e. a set of points of dimensionality less than one. �
��
�� If one runs for a million points one can go reasonably to layer 20, and extract a slope of about 0.5-0.6. The ODE was solved using 13 digits, and one has to worry about the inaccuracies that will accumulate in very long time sequences. From this perspective the measurement of the capcity dimension is problematic with the present method. Various methods have been devised to accelerate the convergence of the capacity dimension. �
��
�� Look at research articles in the journal Physics Letters A: �
�� Phys. Lett. A 141, p. 386; A 148, p. 63; A 151, p.43 have O(N*log(N)) accurate methods, and even O(N) methods apparently exist. �
��
�� Exercise 3: �
�� Experiment with different data sets, in particular, confirm the finding of the small-epsilon regime by using a larger sample of attractor points. �
��
��
�� It is also illustrative to look at histograms over � x � or � y � : �
��
| > | �Nh:=1000: for i from 1 to Nh do: Pxh[i]:=0: od: � |
�
| > | �Nbin:=0: for k from 1 to 200000 do: ix:=trunc(Nh/10*(LL[k][1]+5)); if ix>0 and ix<Nh then if Pxh[ix]=0 then Nbin:=Nbin+1; fi: Pxh[ix]:=Pxh[ix]+1: fi: od: print("Nr of occ. bins: ",Nbin); � |
�
![]() �
�
�
| > | �plot([seq([i,Pxh[i]],i=1..Nh)],style=point,symbol=point,axes=boxed,view=[100..900,0..20]); � |
�
![[Maple Plot]](images/FractalDimension78.gif) �
�
�
| > | �plot([seq([i,Pxh[i]],i=1..Nh)],style=point,symbol=point,axes=boxed,view=[100..900,0..10]); � |
�
![[Maple Plot]](images/FractalDimension79.gif) �
�
�
| > | �Nbin:=0: Nh:=1000: for i from 1 to Nh do: Pyh[i]:=0: od: � |
�
| > | �for k from 1 to 100000 do: iy:=trunc(Nh/10*(LL[k][2]+7)); if iy>0 and iy<Nh then if Pyh[iy]=0 then Nbin:=Nbin+1; fi: Pyh[iy]:=Pyh[iy]+1: fi: od: print("Nr of occ. bins: ",Nbin); � |
�
![]() �
�
�
| > | �plot([seq([i,Pyh[i]],i=1..Nh)],style=point,symbol=point,axes=boxed,view=[100..900,0..20]); � |
�
![[Maple Plot]](images/FractalDimension81.gif) �
�
� The histograms over x or y only, i.e., over position or velocity only are similar in the sense that about 2/3 of the bins are being filled at all, the majority are filled with relatively few points, and a few bins (we find seven; to reveal them: repeat the plots without a y-range restriction!) get filled very strongly. For an object of dimensionality greater than one we should not be missing any bins in the 1d histogram. Thus, it is an interesting question to ask whether there are gaps in the histogram. Will these gaps be filled if more points are added to the Poincare section? �
��
�� Exercise4: �
�� Scan the histogram to find the gaps. What are the regions of x (or y) that are left out? Find out whether they are filled in eventually. �
��
| > | �