� Multidimensional FFT �
��
�� A test of Maple's new discrete Fourier transform package (available as of Maple9). First, we test it in one dimension and demonstrate the effect of two mesh distributions. �
��
�� First case: a one-dimensional mesh that includes the origin, and is similar in both the coordinate and momentum domain. We restrict ourselves to meshes with size in powers of two (as in the old FFT program), even though the algorithm is capable of handling other meshes as well. We will use the interpretation of the Fourier transform as familiar from quantum mechanics, i.e., connecting the coordinate and momentum representations. The alternative would be to look at time signals in the frequency domain. �
�� We do not overemphasize the QM aspect, i.e., we work in units where Planck's constant (hbar) equals unity. Our domains of integration will be symmetric both in � x � and in � p � . �
��
�� 1. One-dimensional FFT �
��
��
| > | �restart; Digits:=15; � |
�
![]() �
�
�
| > | �with(DiscreteTransforms); � |
�
![]() �
�
�
| > | �N:=64; � |
�
![]() �
�
�
| > | �dx:=0.3; � |
�
![]() �
�
�
| > | �dp:=evalhf(2*Pi/(N*dx)); � |
�
![]() �
�
� Our choice of parameters resulted in a similar spacing in coordinate and momentum. We pick a function to be transformed. �
��
| > | �fx:=1/(1+x^2)^2; � |
�
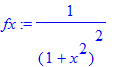 �
�
�
| > | �Z:=Array(1..N,datatype=complex[8]); � |
�
 �
�
� In the first 1d implementation we introduce a coordinate mesh which includes the origin. The mesh is arranged in such a way that one starts with zero, goes to the largest represented positive value, then continues with the largest negative value (which is one unit further than the largest positive) and returns to the origin. �
��
| > | �for i from 1 to N/2 do: � |
�
| > | �pv[i]:=evalhf((i-1)*dp); pv[N-i+1]:=evalhf(-i*dp); xv[i]:=evalhf((i-1)*dx); Z[i]:=eval(fx,x=xv[i]); xv[N-i+1]:=evalhf(-i*dx); Z[N-i+1]:=eval(fx,x=xv[N-i+1]): od: � |
�
| > | �plot([seq([xv[i],Z[i]] ,i=1..N)],x=-10..10,style=point); � |
�
![[Maple Plot]](images/MultidFFT8.gif) �
�
� Note that our mesh spends a lot of points where the function is very small. �
��
�� Now we carry out the Fourier transform whereby the result is stored in the original array: �
��
| > | �FourierTransform(Z,inplace=true); � |
�
 �
�
�
| > | �Z[1],Z[2],Z[N]; � |
�
![]() �
�
� Note the negligible imaginary parts. The discrete FT recognized the symmetry of the function. We graph both real and imaginary parts to confirm this: �
��
| > | �plot([seq([pv[i],Re(Z[i])] ,i=1..N)],style=point); � |
�
![[Maple Plot]](images/MultidFFT11.gif) �
�
�
| > | �plot([seq([pv[i],Im(Z[i])] ,i=1..N)],style=point); � |
�
![[Maple Plot]](images/MultidFFT12.gif) �
�
� How well does the DFT agree with the known answer? �
��
| > | �fp:=int(fx*exp(I*p*x),x=-infinity..infinity) assuming p,real; � |
�
![]() �
�
�
| > | �evalf(limit(fp,p=0)); � |
�
![]() �
�
�
| > | �plot(fp,p=-10..10); � |
�
![[Maple Plot]](images/MultidFFT15.gif) �
�
�
| > | �Z[1]; � |
�
![]() �
�
� Before we can compare in detail we have to resolve some normalization issues. Neither our integral, nor the Maple DFT contain the customary � 1/sqrt(2*Pi) � factor. Maple's DFT contains a � 1/sqrt(N) � factor, but is missing a � dx � factor to form the Riemann sum (which connects the result to the integral). Thus, we correct for both: �
��
| > | �f_DFT:=evalf(sqrt(N)*dx): f_DFT*Z[1]; � |
�
![]() �
�
�
| > | �P1:=plot([seq([pv[i],Re(f_DFT*Z[i])] ,i=1..N)],style=point): � |
�
| > | �P2:=plot(fp,p=-10..10,color=blue): plots[display](P1,P2); � |
�
![[Maple Plot]](images/MultidFFT18.gif) �
�
� The agreement is very good. �
��
�� Exercise 1.1: �
�� Explore the question of agreement/disagreement in detail: calculate the percentage error and display it for all momenta ( � p � -values). �
��
�� Exercise 1.2: �
�� Explore the behaviour using different values of � N, dx: � can one obtain acceptable results with small � N � ? �
��
�� Exercise 1.3: �
�� Modify the function pair � {fx, fp} � (you can replace the � 1 � in the denominator by a constant), or change to a different pair (different power of the entire denominator?) and explore Exercises 1 and 2. �
��
�� Mini-project: � �
�� Explore further issues with the FFT: calculate the second derivate of the function by going to transform space, multiplying with � p^2 � , and transforming back using � InverseFourierTransform � , and compare with the analytical result. �
��
| > | �
� Now we try a modification of the mesh that excludes the origin in the � x � -domain. This is useful for cases when the original function is undefined there. �
��
| > | �
�
| > | �restart; Digits:=15: � |
�
| > | �with(DiscreteTransforms); � |
�
![]() �
�
�
| > | �N:=64; � |
�
![]() �
�
�
| > | �Z:=Array(1..N,datatype=complex[8]); � |
�
 �
�
�
| > | �dx:=0.3; � |
�
![]() �
�
�
| > | �xM:=Vector(N): � |
�
| > | �for i from 1 to N/2 do: xM[i]:=(i-1/2)*dx; xM[N-i+1]:=-xM[i]; od: � |
�
| > | �xM[1],xM[N/2],xM[N/2+1],xM[N]; � |
�
![]() �
�
�
| > | �dpx:=evalf(2*Pi/(N*dx)); � |
�
![]() �
�
� It will turn out that the transformed variable domain does contain the � p=0 � point: �
��
| > | �for i from 1 to N/2+1 do: pxM[i]:=(i-1)*dpx: � |
�
| > | �if i>1 then pxM[N-i+2]:=-pxM[i]: fi: od: � |
�
| > | �pxM[1],pxM[2],pxM[N/2],pxM[N/2+1],pxM[N]; � |
�
![]() �
�
�
| > | �
�
| > | �a:=1; � |
�
![]() �
�
�
| > | �fx:=1/(a^2+x^2)^2; � |
�
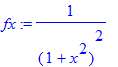 �
�
�
| > | �fp:=int(fx*exp(I*p*x),x=-infinity..infinity) assuming p,real; � |
�
![]() �
�
�
| > | �for i from 1 to N do: x:=xM[i]: � |
�
| > | �Z[i]:=fx: od: x:='x': � |
�
| > | �FourierTransform(Z,inplace=true); � |
�
 �
�
�
| > | �Z[1],Z[2]; � |
�
![]() �
�
� Why is this complex-valued? �
�� Arrange the density in an array with p ranging from negative to positive. �
��
| > | �plot([seq([pxM[i],Re(Z[i])],i=1..N)],style=point); � |
�
![[Maple Plot]](images/MultidFFT31.gif) �
�
�
| > | �plot([seq([pxM[i],Im(Z[i])],i=1..N)],style=point); � |
�
![[Maple Plot]](images/MultidFFT32.gif) �
�
�
| > | �f_DFT:=evalf(sqrt(N)*dx); � |
�
![]() �
�
�
| > | �P1:=plot([seq([pxM[i],Re(f_DFT*Z[i])],i=1..N)],style=point): � |
�
| > | �P2:=plot(fp,p=-10..10,color=blue): plots[display](P1,P2); � |
�
![[Maple Plot]](images/MultidFFT34.gif) �
�
� As could have been guessed, the real part alone does not agree with the answer. If we take the absolute value, matters are corrected. Thus, for this mesh arrangement we obtain a DFT which contains a wrong complex phase. �
��
| > | �P1:=plot([seq([pxM[i],abs(f_DFT*Z[i])],i=1..N)],style=point): � |
�
| > | �P2:=plot(fp,p=-10..10,color=blue): plots[display](P1,P2); � |
�
![[Maple Plot]](images/MultidFFT35.gif) �
�
� Therefore, it might be better to work with the mesh that contains the origin, and to use a regulator in those instances where the function to be transformed has problems at the origin. �
��
�� Exercise 1.4: �
�� Explore the effect of the phase error by reducing � dx � . You can increase � N � or keep it constant. What happens to the real and imaginary parts of the DFT? �
��
�� Exercise 1.5: �
�� An interesting example to Fourier analyze is the Gaussian wavepacket which one can control through various parameters such as width � Dx � (coordinate space parameter corresponds to inverse momentum width parameter), and center momentum � p0 � . It is relatively straightforward to find parameter choices which exceed the mesh capabilities, either by exceeding the momentum resolution (very broad packet in � x � -space = narrow structure in � p � -space), or by putting � p0 � at the edge of the covered momentum range. Explore this on your own. � GWP=Norm*exp(-(x/Dx)^2)*exp(I*p0*x) � . Find the analytic Fourier image and compare with the FFT result. �
��
| > | �
� 2. Two-dimensional FFT �
��
�� Now we proceed with the two-dimensional case. Comparison with analytical results will be more complicated. In contrast to the old FFT routines multi-dimensional FFTs are calculated within the � DiscreteTransforms � package. �
��
| > | �restart; Digits:=15: with(DiscreteTransforms); � |
�
![]() �
�
�
| > | �N:=128; � |
�
![]() �
�
�
| > | �Z:=Array(1..N,1..N,datatype=complex[8]); � |
�
 �
�
�
| > | �dx:=0.3; dy:=dx: � |
�
![]() �
�
� The simplest function to choose would be one which factorizes. �
��
| > | �a:=1; � |
�
![]() �
�
�
| > | �fx:=1/(a^2+x^2)^3; � |
�
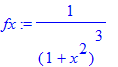 �
�
�
| > | �fpx:=int(fx*exp(I*px*x),x=-infinity..infinity) assuming px,real; � |
�
![]() �
�
![]() �
�
�
| > | �b:=2; � |
�
![]() �
�
�
| > | �fy:=1/(b^2+y^2)^3; � |
�
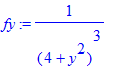 �
�
�
| > | �fpy:=int(fy*exp(I*py*y),y=-infinity..infinity) assuming py,real; � |
�
![]() �
�
![]() �
�
�
| > | �
�
| > | �xM:=Vector(N): yM:=Vector(N): � |
�
| > | �for i from 1 to N/2 do: xM[i]:=(i-1/2)*dx; xM[N-i+1]:=-xM[i]; yM[i]:=(i-1/2)*dy; yM[N-i+1]:=-yM[i]; od: � |
�
| > | �xM[1],xM[N/2],xM[N/2+1],xM[N],yM[1],yM[N]; � |
�
![]() �
�
� We chose a cube, but nothing prevented us from using a parallelepiped as a computational domain. Simply use different dy, dz. �
��
| > | �for i from 1 to N do: x:=xM[i]: for j from 1 to N do: y:=yM[j]: � |
�
| > | �r:=sqrt(x^2+y^2): Z[i,j]:=fx*fy: od: od: � |
�
| > | �DA:=Array(1..N,1..N,1..3,datatype=float[8]): � |
�
| > | �#for i from 1 to N do: x:=xM[i]: for j from 1 to N do: y:=yM[j]: DA[i,j,1]:=x: DA[i,j,2]:=y: DA[i,j,3]:=Z[i,j]; od: od: � |
�
| > | �for i from 1 to N do: x:=xM[i]: for j from 1 to N do: y:=yM[j]: DA[i,j,1]:=x: DA[i,j,2]:=y: DA[i,j,3]:=log(abs(Z[i,j])); od: od: � |
� The � surfdata � procedure is terrible at plotting the original surface. That's why we graph the log. �
��
| > | �plots[surfdata](DA,axes=boxed,style=patchcontour,shading=zhue); � |
�
![[Maple Plot]](images/MultidFFT49.gif) �
�
�
| > | �FourierTransform(Z,inplace=true); � |
�
 �
�
�
| > | �Z[1,2]; � |
�
![]() �
�
� Why is this complex-valued? The one-dimensional results imply that the mesh distribution without an origin in the original � r � -domain are to blame. �
��
| > | �dpx:=evalf(2*Pi/(N*dx)); � |
�
| > | �dpy:=evalf(2*Pi/(N*dy)): � |
�
![]() �
�
�
| > | �for i from 1 to N/2+1 do: pxM[i]:=(i-1)*dpx: pyM[i]:=(i-1)*dpy: � |
�
| > | �if i>1 then pxM[N-i+2]:=-pxM[i]: pyM[N-i+2]:=-pyM[i]: fi: od: � |
�
| > | �pxM[1],pxM[2],pxM[N/2],pxM[N/2+1],pxM[N]; � |
�
![]() �
�
� At one point we were so dismayed by the bad surface plot that we exported the data, and used Surfer 7 to graph them. The grid generation and contour plot in that program is much superior to � surfdata � . We left the commented commands to show how to export data via ASCII files. �
��
| > | �# fd := fopen("C:/marko/maple9/testsrf.dat",WRITE); � |
�
| > | �DA:=Array(1..N,1..N,1..3,datatype=float[8]): � |
�
| > | �for i from 1 to N do: px:=pxM[i]: for j from 1 to N do: py:=pyM[j]: DA[i,j,1]:=px: DA[i,j,2]:=py: DA[i,j,3]:=abs(Z[i,j]); od: od: � |
�
| > | �px:='px': py:='py': � |
�
| > | �# fprintf(fd,"%g %g %g\n",px,py,DA[i,j,3]); � |
�
| > | �for i from 1 to N do: for j from 1 to N do: if DA[i,j,3]>0 then DA[i,j,3]:=log10(DA[i,j,3]): else DA[i,j,3]:=0: fi: od: od: � |
� We cheat here: the zero values that occur at the largest momentum value are transferred to the origin, where we enter the known value. Didn't know how else to get rid off the value. �
��
| > | �for i from 1 to N do: for j from 1 to N do: if DA[i,j,3]=0 then DA[i,j,1]:=0: DA[i,j,2]:=0; DA[i,j,3]:=DA[1,1,3]; fi: od: od: � |
�
| > | �# fclose(fd); � |
�
| > | �plots[surfdata](DA,axes=boxed,style=patchcontour,shading=zhue); � |
�
![[Maple Plot]](images/MultidFFT54.gif) �
�
� Why the surface looks different viewed from the top versus the bottom is for the � surfdata � programmers to explain. �
�� There are cases when the graph looks bad from the top, but has OK contours when looked at from below. � FT(exp(-r)) � is an example. �
��
�� How do we know the calculated 2dimensional FT is correct? We would have to use our known FT pair and plug in the correct conversion factor. �
��
| > | �f_DFT2:=evalf(sqrt(N)*dx*sqrt(N)*dy); � |
�
![]() �
�
�
| > | �Z[1,1]*f_DFT2; � |
�
![]() �
�
�
| > | �evalf(eval(fpx*fpy,{x=0,y=0})); � |
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
�
| > | �plot3d(log10(abs(fpx*fpy/f_DFT2)),px=-10..10,py=-10..10,axes=boxed,style=patchcontour,shading=zhue); � |
�
![[Maple Plot]](images/MultidFFT62.gif) �
�
� This looks OK what about the details?. �
��
��
| > | �n_x:=15; n_y:=10; � |
�
![]() �
�
�
![]() �
�
�
| > | �DA[n_x,n_y]; � |
�
![]() �
�
�
| > | �evalf(eval(log10(abs(fpx*fpy/f_DFT2)),{px=pxM[n_x],py=pyM[n_y]})); � |
�
![]() �
�
� The agreement at intermediate momentum values is excellent! �
��
�� Exercise 2.1: �
�� Find other FT function pairs (ideally non-factorized ones) and observe the accuracy of the DFT. �
��
| > | �
� 3. Three-dimensional FFT �
��
| > | �restart; Digits:=15: with(DiscreteTransforms); � |
�
![]() �
�
�
| > | �N:=64; � |
�
![]() �
�
�
| > | �Z:=Array(1..N,1..N,1..N,datatype=complex[8]); � |
�
 �
�
�
| > | �dx:=0.3; dy:=dx: dz:=dy: � |
�
![]() �
�
�
| > | �xM:=Vector(N): yM:=Vector(N): zM:=Vector(N): � |
�
| > | �for i from 1 to N/2 do: xM[i]:=(i-1/2)*dx; xM[N-i+1]:=-xM[i]; yM[i]:=(i-1/2)*dy; yM[N-i+1]:=-yM[i]; zM[i]:=(i-1/2)*dz; zM[N-i+1]:=-zM[i]; od: � |
�
| > | �xM[1],xM[N],yM[1],yM[N],zM[1],zM[N]; � |
�
![]() �
�
� We chose a cube, but nothing prevented us from using a parallelepiped as a computational domain. Simply use different � dy, dz � . �
��
| > | �a:=1; � |
�
![]() �
�
�
| > | �for i from 1 to N do: x:=xM[i]: for j from 1 to N do: y:=yM[j]: for k from 1 to N do: z:=zM[k]: � |
�
| > | �r:=sqrt(x^2+y^2+z^2): Z[i,j,k]:=exp(-a*r): od: od: od: � |
�
| > | �FourierTransform(Z,inplace=true); � |
�
 �
�
�
| > | �Z[1,1,2]; � |
�
![]() �
�
� Why is this complex-valued? �
��
| > | �dpx:=evalf(2*Pi/(N*dx)); � |
�
![]() �
�
�
| > | �dpy:=evalf(2*Pi/(N*dy)): dpz:=evalf(2*Pi/(N*dz)): � |
�
| > | �for i from 1 to N/2+1 do: pxM[i]:=(i-1)*dpx: pyM[i]:=(i-1)*dpy: pzM[i]:=(i-1)*dpz: � |
�
| > | �if i>1 then pxM[N-i+2]:=-pxM[i]: pyM[N-i+2]:=-pyM[i]: pzM[N-i+2]:=-pzM[i]: fi: od: � |
�
| > | �pxM[1],pxM[2],pxM[N/2],pxM[N/2+1],pxM[N]; � |
�
![]() �
�
� Now look at a cut. Arrange the density in an array with p ranging from negative to positive. �
��
| > | �DA:=Array(1..N,1..N,1..3,datatype=float[8]): � |
�
| > | �for i from 1 to N do: px:=pxM[i]: for j from 1 to N do: py:=pyM[j]: DA[i,j,1]:=px: DA[i,j,2]:=py: DA[i,j,3]:=0: for k from 1 to N do: pz:=pzM[k]: DA[i,j,3]:=DA[i,j,3]+abs(Z[i,j,k])*dpz; od: od: od: � |
�
| > | �for i from 1 to N do: for j from 1 to N do: if DA[i,j,3]>0 then DA[i,j,3]:=log10(DA[i,j,3]): else DA[i,j,3]:=0: fi: od: od: � |
�
| > | �for i from 1 to N do: for j from 1 to N do: if DA[i,j,3]=0 then DA[i,j,3]:=DA[1,1,3]: DA[i,j,1]:=DA[1,1,1]: DA[i,j,2]:=DA[1,1,2]: fi: od: od: � |
�
| > | �plots[surfdata](DA,axes=boxed,style=patchcontour,shading=zhue); � |
�
![[Maple Plot]](images/MultidFFT77.gif) �
�
� This is a case that looks better from the bottom than from above. �
��
| > | �f_DFT3:=evalf(sqrt(N)*dx*sqrt(N)*dy*sqrt(N)*dz); � |
�
![]() �
�
� How can we verify the validity? We need to calculate the analytic FT for our function. This is done in spherical polar coordinates. The azimuthal angular integral yields � 2*Pi � . The polar angle integration is straightforward. The radial integral is done in Maple: �
��
| > | �r:='r': fp:=simplify(4*Pi/p*int(exp(-a*r)*sin(p*r)*r,r=0..infinity)) assuming p>0; � |
�
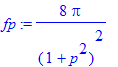 �
�
� We have a spherically symmetric answer. We should be able to verify the FT result by choosing a line along a beam starting at � p=0 � . �
��
| > | �ResN:=Vector(N): ResA:=Vector(N): � |
�
| > | �for i from 1 to N/2 do: px:=pxM[i]: py:=pyM[i]: pz:=pzM[i]: p:=sqrt(px^2+py^2+pz^2): ResA[i]:=[p,p*evalf(fp)]: ResN[i]:=[p,p*abs(f_DFT3*Z[i,i,i])]: od: � |
�
| > | �ResA[2],ResN[2]; � |
�
![]() �
�
�
| > | �P1:=plot([seq(ResN[i],i=1..N/2)],style=point): � |
�
| > | �P2:=plot([seq(ResA[i],i=1..N/2)],color=blue): plots[display](P1,P2); � |
�
![[Maple Plot]](images/MultidFFT81.gif) �
�
� The FFT works remarkably well given that we have not resolved the maximum in the � p*f(p) � distribution, i.e., we are working on a relatively coarse mesh. �
�� Keep in mind that we used the absolute value of the transform. A mesh including the origin should not suffer from this problem. �
��
�� Exercise 2.2: �
�� Change the parameter � a � in the wavefunction and observe the change in the momentum distribution. Explain what it means physically. Find other function pairs (separable in Cartesians, and non-separable) and verify how well the FFT works. �
��
�� Exercise 2.3: �
�� For QM-literate students: consider the calculation of the kinetic energy expectation value in momentum space. You can calculate it by multiplying in � p � -space with the multiplicative operator � p^2/2 � (in our units � m=hbar=1 � ) and forming the average (and normalizing properly). This will emphasize larger � p � -values. To check how accurate this calculation will be you can probe the agreement of � p^2*f(p) � in the above plot. What do you find? How can you improve upon the situation? �
��
�� Exercise 2.4: �
�� Change the mesh in the above code to include the origin also in the configuration space � (x,y,z) � domain. Demonstrate that the problems with the incorrect complex phase disappear in this case. �
��
�� Mini-project: �
�� Demonstrate the usefulness of the 3d FFT by making other cuts for comparison between numerical and analytical results. �
��
| > | �