� Discrete Wigner Function �
��
�� We use the discrete Fourier transform to carry out Wigner transforms of wavepackets that represent more complicated situations than those available by means of symbolic integration, and explored in the worksheet � Wigner.mws � . We are interested, in particular, in the Wigner distribution for a tunneling wavepacket as calculated by numerical techniques. First, we test the calculation on a simple Gaussian wavepacket for which the analytic answer is available. �
��
��
| > | �restart; a:=1; p0:='p0'; � |
�
![]() �
�
�
![]() �
�
�
| > | �psi := unapply(sqrt(a)*1/Pi^(1/4)*exp(-1/2*(x-I*a^2*p0)^2/(a^2+t*I)-1/2*a^2*p0^2)/(sqrt(-a^2-I*t)),x); � |
�
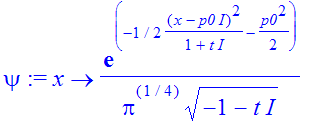 �
�
�
| > | �assume(t>0,p,real); � |
�
| > | �int(evalc(simplify(abs(psi(x))^2)),x=-infinity..infinity); � |
�
![]() �
�
�
| > | �rho:=simplify(expand(psi(x+s/2)*conjugate(psi(x-s/2)))); � |
�
![]() �
�
![]() �
�
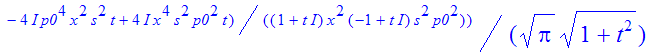 �
�
� Our task is now to perform a discrete FT over the variable � s � to map it into a momentum � p � . This is to be carried out for a number of � x � values, and in this way a distribution over momenta � p � and positions � x � will be obtained. Of course, we have to pick numerical values for the average momentum and for time. �
��
| > | �p0:=1; � |
�
![]() �
�
�
| > | �rho2:=simplify(subs(t=2,rho)); � |
�
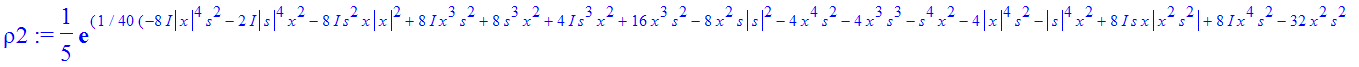 �
�
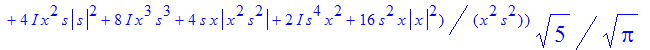 �
�
� We borrow the code from the quantum mechancis example at the end of the worksheet � FFT.mws � to generate the mesh, and to carry out the DFT. We re-label � x � to � s � in the code in order to avoid confusion with the current use of � x � as the position in the Wigner distribution argument. We are particularly interested in small-size FT explorations. The idea is that small � s � -meshes centered around the actual value of � x � should suffice. This becomes an important issue for multi-dimensional calculations (beyond the scope of Maple, but doable in Fortran). The reason why we have to be careful is that the DFT assumes periodicity, and a small mesh can mean that we work with an artificial modification of the density matrix � rho � . �
��
| > | �ds:=0.8; n:=5; N:=2^n; � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �dp:=evalhf(2*Pi/(N*ds)); � |
�
![]() �
�
�
| > | �p_max:=N*dp/2; � |
�
![]() �
�
�
| > | �sv:=array(1..N): pv:=array(1..N): � |
�
| > | �for i from 1 to N do: � if i < N/2+1 then � sv[i]:=evalhf((i-1)*ds): sv[N-i+1]:=evalhf(-i*ds): � pv[i]:=evalhf((i-1)*dp): pv[N-i+1]:=evalhf(-i*dp): end if: � od: � |
� Let us try one � x � value first: �
��
| > | �xval:=2; � |
�
![]() �
�
� A small technical wrinkle: for � s=0 � (the first value, i.e., for � sv[1] � ) the expression for the density matrix is undefined (0/0); therefore, we don't substitute � s=sv[i] � , but rather take the limit. To figure this out required a little debugging step. �
��
| > | �yR:=array(1..N): yI:=array(1..N): � |
�
| > | �for i from 1 to N do: yR[i]:=evalf(limit(subs(x=xval,Re(rho2)),s=sv[i])); yI[i]:=evalf(limit(subs(x=xval,Im(rho2)),s=sv[i])); od: � |
�
| > | �plot([seq([sv[i],yR[i]],i=1..N)],style=point); � |
�
![[Maple Plot]](images/WignerDiscrete17.gif) �
�
�
| > | �plot([seq([sv[i],yI[i]],i=1..N)],style=point,color=blue); � |
�
![[Maple Plot]](images/WignerDiscrete18.gif) �
�
�
| > | �N_s:=ds*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
![]() �
�
�
| > | �evalhf(FFT(n, var(yR), var(yI))); � |
�
![]() �
�
�
| > | �N_p:=dp*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
![]() �
�
� The imaginary part should vanish; is this true? �
��
| > | �dp*add(yI[i]^2,i=1..N); � |
�
![]() �
�
� Well, almost. Perhaps this represents a good test of the accuracy of our truncation of the transform to a low number of points. �
��
| > | �yRn:=array(1..N): sq:=evalf(1/sqrt(2*Pi)): for i from 1 to N do: yRn[i]:=ds*sq*yR[i]: od: � |
�
| > | �N_p:=dp*add(yRn[i]^2,i=1..N); � |
�
![]() �
�
�
| > | �plot([seq([pv[i],yRn[i]],i=1..N)],style=line); � |
�
![[Maple Plot]](images/WignerDiscrete24.gif) �
�
� Now we set up a loop to step through � x � : �
��
| > | �Tnorm:=0: for k from 1 to 25 do: xval:=evalf(-4+8/20*(k-0.5)); � |
�
| > | �yR:=array(1..N): yI:=array(1..N): � |
�
| > | �for i from 1 to N do: yR[i]:=evalf(limit(subs(x=xval,Re(rho2)),s=sv[i])); yI[i]:=evalf(limit(subs(x=xval,Im(rho2)),s=sv[i])); od: � |
�
| > | �N_s:=ds*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
| > | �evalhf(FFT(n, var(yR), var(yI))); � |
�
| > | �yRn:=array(1..N): sq:=evalf(1/sqrt(2*Pi)): for i from 1 to N do: yRn[i]:=ds*sq*yR[i]: od: � |
�
| > | �N_p:=dp*add(yRn[i]^2,i=1..N); print("xval,Ns,Np=",xval,N_s,N_p); Tnorm:=Tnorm+8/20*N_p; � |
�
| > | �PL[k]:=plot([seq([pv[i],yRn[i]],i=1..N)],style=line,title=cat("x= ",trunc(10*xval),"/10")): od: Tnorm; � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �plots[display]([seq(PL[i],i=1..Nk)],insequence=true); � |
� We observe that the Wigner distribution resembles the analytical result displayed as a surface plot over x and p. It is positive at all values displayed (as it should be for a Gaussian wavepacket). The total norm preservation is very good. The use of the FFT with few points results in a coarse mesh of momentum values. �
��
�� Exercise 1: �
�� Explore the parameter choices for the FFT that give reasonable results. What is more important: a fine resolution in � s � , or a sufficient range? �
��
�� In order to display the results for a wavepacket with tunneling we first have to construct a wavepacket with such properties. One alternative would be to use a FFT-based propagation scheme as introduced in � Ehrenfest.mws � . This integrator works for arbitrary potentials. For piecewise constant potentials we can use a plane-wave decomposition of the wavepacket. Let us develop this approach. While it is rather restrictive in applicability, it offers a link between what is usually done in quantum mechancis texts, namely the stationary scattering theory approach with the time-dependent wavepacket approach. �
��
�� We assume that the potential barrier is different from zero only in the range � 0 < x < 1 � , and that it has height � V0 � there, and is zero everywhere else. For an asymptotic wavenumber value � k0 � we can then calculate the plane wave state. The particle mass is chosen to be � m=1 � in our units. We have to construct solutions to the Schroedinger equation from plane-wave segments, i.e., we calculate energy eigenstates. The scattering is assumed to proceed from left to right. For � x<0 � for a given energy there can be a right-travelling, and a left-travelling plane wave (the latter describing the backscattered flux). For � 0 < x < 1 � we need a superposition, and for � x>1 � we need only a right-travelling wave. The waves have to be matched with continuous derivative at the two points where the potential changes in a step-like fashion. �
��
| > | �
�
| > | �restart; � |
�
| > | �V0:=2; � |
�
![]() �
�
�
| > | �q0:=k0->sqrt(k0^2-2*V0); � |
�
![]() �
�
�
| > | �PW:=k->piecewise(x<0,exp(I*k*x)+ R*exp(-I*k*x), x>=0 and x<=1,A*exp(I*q0(k)*x) + B*exp(-I*q0(k)*x), x>1, T*exp(I*k*x)); � |
�
![]() �
�
� In order to find an acceptable solution for a given energy � E = k^2/2 � we need to determine the complex-valued constants � R, T, A, B � . �
�� The squared magnitudes of R and T correspond to the reflection and transmission probabilities, because the incident flux has been normalized to unity (amplitude of 1 for the right-travelling wave at � x<0 � ). �
��
| > | �k0:=1.; � |
�
![]() �
�
�
| > | �E0:=k0^2/2; � |
�
![]() �
�
� We are testing at an energy which corresponds to one half of the barrier height. Our first attempt to formulate the condition at � x=0 � reveals a � bug in maple � 8. There should be no � x � left in the expression after taking the limit. We should be able to substitute � x=0 � , however, given that it is included in the piecewise definition. �
��
��
| > | �C1:=limit(PW(k0),x=0,left)=simplify(limit(PW(k0),x=0,right)); � |
�
![]() �
�
�
| > | �C1:=limit(PW(k0),x=0,left)=simplify(subs(x=0,PW(k0))); � |
�
![]() �
�
�
| > | �C2:=limit(PW(k0),x=1,right)=simplify(subs(x=1,PW(k0))); � |
�
![]() �
�
![]() �
�
![]() �
�
� Our patience with Maple's � piecewise � function has reached its limit! Maple developers have to return to the drawing board. �
�� Let us see whether another construct available in Maple will help. Let us define explicitly: �
��
��
| > | �PW:=k->if x<0 then exp(I*k*x)+ R*exp(-I*k*x) elif x>=0 and x<=1 then A*exp(I*q0(k)*x) + B*exp(-I*q0(k)*x) else T*exp(I*k*x) fi; � |
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
![]() �
�
�
| > | �C1:=limit(PW(k0),x=0,left)=simplify(limit(PW(k0),x=0,right)); � |
�
Error, (in PW) cannot determine if this expression is true or false: x < 0
�
�
� Ouch! So let us simply define the three adjacent pieces as separate expressions. We label them as parts 1, 2, 3 for the regions � x<0 � , � 0<x<1 � , and � x>1 � . �
��
| > | �PW1:=k->exp(I*k*x)+ R*exp(-I*k*x); � |
�
![]() �
�
�
| > | �PW2:=k->A*exp(I*q0(k)*x) + B*exp(-I*q0(k)*x); � |
�
![]() �
�
�
| > | �PW3:=k->T*exp(I*k*x); � |
�
![]() �
�
� This allows us to avoid the contentious issue in Maple of taking the limit on a piecewise function. For graphing we will want a single function later. �
��
| > | �PW:=k->piecewise(x<0,PW1(k), x>=0 and x<=1,PW2(k), x>1, PW3(k)); � |
�
![]() �
�
�
| > | �C1:=simplify(subs(x=0,PW1(k0)))=simplify(subs(x=0,PW2(k0))); � |
�
![]() �
�
�
| > | �C2:=simplify(subs(x=1,PW2(k0)))=simplify(subs(x=1,PW3(k0))); � |
�
![]() �
�
� Define the derivatives: �
��
| > | �PW1p:=unapply(simplify(diff(PW1(k),x)),k); � |
�
![]() �
�
�
| > | �PW2p:=unapply(simplify(diff(PW2(k),x)),k); � |
�
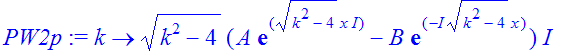 �
�
�
| > | �PW3p:=unapply(simplify(diff(PW3(k),x)),k); � |
�
![]() �
�
� Formulate the continuity of the derivative: �
��
| > | �C1p:=simplify(subs(x=0,PW1p(k0)))=simplify(subs(x=0,PW2p(k0))); � |
�
![]() �
�
�
| > | �C2p:=simplify(subs(x=1,PW2p(k0)))=simplify(subs(x=1,PW3p(k0))); � |
�
![]() �
�
�
| > | �sol:=solve({C1,C2,C1p,C2p},{A,B,T,R}); � |
�
![]() �
�
![]() �
�
� The solution works when � k0 � is entered as a floating point number. For some reason it fails in exact mode for � k0=1 � . �
�� To use the coefficients we assign the solution. �
��
| > | �assign(sol); � |
�
| > | �plot([Re(PW(k0)),Im(PW(k0))],x=-10..10,color=[red,blue]); � |
�
![[Maple Plot]](images/WignerDiscrete81.gif) �
�
�
| > | �PW(k0); � |
�
![PIECEWISE([exp(1.*I*x)-(.4545165750+.8381216098*I)*exp(-1.*I*x), x < 0],[(.5146865809-.8389435728*I)*exp(-1.732050808*x)+(.3079684405e-1+.8219630811e-3*I)*exp(1.732050808*x), -x <= 0 and x-1 <= 0],[(.2...](images/WignerDiscrete82.gif) �
�
� Now that we know how this works, let us assemble solutions for a range of wavenumber values: �
��
| > | �k0:=0.01: k_max:=2.5: N_k:=80: dk:=evalf((k_max-k0)/N_k); � |
�
![]() �
�
�
| > | �for i_k from 1 to N_k do: k_i:=evalf(k0+(i_k-1)*dk); � |
�
| > | �A:='A': B:='B': R:='R': T:='T': � |
�
| > | �C1:=simplify(subs(x=0,PW1(k_i)))=simplify(subs(x=0,PW2(k_i))); � |
�
| > | �C2:=simplify(subs(x=1,PW2(k_i)))=simplify(subs(x=1,PW3(k_i))); � |
�
| > | �C1p:=simplify(subs(x=0,PW1p(k_i)))=simplify(subs(x=0,PW2p(k_i))); � |
�
| > | �C2p:=simplify(subs(x=1,PW2p(k_i)))=simplify(subs(x=1,PW3p(k_i))); � |
�
| > | �sol:=solve({C1,C2,C1p,C2p},{A,B,T,R}); � |
�
| > | �assign(sol): kv[i_k]:=k_i: WF[i_k]:=PW(k_i): od: � |
� We can graph an animation of the basis state solutions: �
��
| > | �#PL:=[seq(plot([Re(WF[i]),Im(WF[i])],x=-5..5,color=[red,blue]),i=1..N_k)]: � |
�
| > | �#plots[display](PL,insequence=true); � |
� Now superimpose those plane-wave scattering solutions: �
��
| > | �#IP:=(f,g)->evalf[6](Int(simplify(evalc(conjugate(f)*g)),x=-infinity..0,method=_Gquad))+evalf[6](Int(simplify(evalc(conjugate(f)*g)),x=0..1,method=_Gquad))+evalf[6](Int(simplify(evalc(conjugate(f)*g)),x=1..infinity,method=_Gquad)); � |
� The above definition leads to slow calculation, therefore, we use a simpler method: �
�� The real axis is discretized and truncated for the purpose of calculating integrals. �
��
| > | �x_min:=-35: x_max:=35: N_i:=200: dx:=evalf((x_max-x_min)/N_i); � |
�
![]() �
�
�
| > | �IP:=(f,g)->dx*add(evalf(subs(x=x_min+(i-0.5)*dx,conjugate(f)*g)),i=1..N_i); � |
�
![]() �
�
�
| > | �a:=3; � |
�
![]() �
�
�
| > | �phi0:=(2/Pi)^(1/4)/sqrt(a)*exp(-(x+10)^2/a^2)*exp(1.*I*x); � |
�
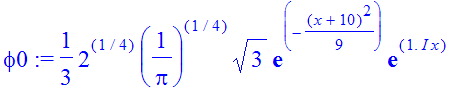 �
�
�
| > | �int(simplify(evalc(abs(phi0)^2)),x=-infinity..infinity); � |
�
![]() �
�
�
| > | �IP(phi0,phi0); � |
�
![]() �
�
� On the other hand, the initial state is completely localized in the � x<0 � region, so we should be able to get away with one half of the work: �
��
| > | �IP0:=(f,g)->dx*add(evalf(subs(x=x_min+(i-0.5)*dx,conjugate(f)*g)),i=1..N_i/2); � |
�
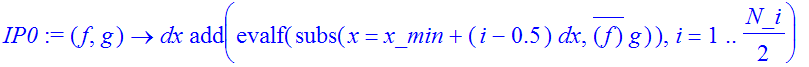 �
�
�
| > | �IP0(phi0,phi0); � |
�
![]() �
�
�
| > | �c_n:=[seq(IP0(WF[n],phi0),n=1..N_k)]: � |
�
| > | �plot([seq([k,abs(c_n[k])^2],k=1..N_k)],style=point); � |
�
![[Maple Plot]](images/WignerDiscrete92.gif) �
�
� The above result us smooth. For different initial wavepackets (e.g., narrower ones, which have a bigger momentum spread) the mesh of wavenumber values included may require modification so that the above collection of coefficients indicates both a reasonable resolution (almost 'smooth' curve), and that the magnitude of the highest coefficients approaches zero. �
��
�� We form a superposition quite naively: for a discrete set of scattering states chosen for some arbitrary set of wavenumber values we do not have a guarantee of orthogonality. In the limit of an infinite real axis these states acquire delta-function normalization. We will have to normalize the wavepacket represented in terms of the scattering states later manually. �
��
| > | �psi:=add(c_n[i]*(dk)*WF[i]*exp(-I*0.5*kv[i]^2*t),i=1..N_k): � |
� The following calculation allows to visualize the time evolution of the wavepacket. It takes some time to compute. �
��
| > | �PLt:=[seq(plot(evalc(abs(subs(t=1.0*i_t,psi))^2),x=-15..10,title=cat("t= ",i_t)),i_t=0..30)]: � |
�
| > | �plots[display](PLt,insequence=true); � |
�
![[Maple Plot]](images/WignerDiscrete93.gif) �
�
�
| > | �IP(subs(t=0,psi),subs(t=0,psi)); � |
�
| > | �IP(subs(t=10,psi),subs(t=10,psi)); � |
�
| > | �No:=1/sqrt(Re(%)); � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
� This normalization constant is used below to have a properly normalized wavepacket. �
��
�� Now we implement the Wigner distribution calculation. The wavepacket is ready for calculation at any reasonable time � t � . First, however, we test the Wigner function calculation by using the Gaussian initial state. �
��
| > | �#phi:=No*subs(t=10,psi): � |
�
| > | �phi:=phi0: � |
� Now map the wavefunction to a mesh on which the Fourier transforms can be carried out. First define a Fourier mesh by lifting code from the above section. The next line serves only as a reminder to explain the relationship between � x � and the Fourier transform variable � s � . �
��
| > | �#rho:=expand(subs(x=x+s/2,phi)*conjugate(subs(x=x-s/2,phi))): � |
�
| > | �ds:=0.8; n:=6; N:=2^n; � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �dp:=evalhf(2*Pi/(N*ds)); � |
�
![]() �
�
�
| > | �p_max:=N*dp/2; � |
�
![]() �
�
�
| > | �sv:=array(1..N): pv:=array(1..N): � |
�
| > | �for i from 1 to N do: � if i < N/2+1 then � sv[i]:=evalhf((i-1)*ds): sv[N-i+1]:=evalhf(-i*ds): � pv[i]:=evalhf((i-1)*dp): pv[N-i+1]:=evalhf(-i*dp): end if: � od: � |
� In order to be able to carry out Wigner distribution calculations we need the wavefunction on a mesh of spacing � dx=0.4 � , chosen to be commensurate with the above Fourier transform mesh. This means that the choice of � x � -values will be restricted. This mesh has to be sufficiently large so that for all points � x � of interest the Wigner transform can be carried out by the � s � -variable while exploring the neighbourhood of � x � . �
��
| > | �N_x:=512: dx:=0.4: xv:=array(1..N_x): phi_x:=array(1..N_x): for i from 1 to N_x do: xv[i]:=(i-N_x/2)*dx: phi_x[i]:=evalf(subs(x=xv[i],phi)): od: � |
�
| > | �PL1:=plot([seq([xv[i],Re(phi_x[i])],i=1..N_x)],color=red): � |
�
| > | �PL2:=plot([seq([xv[i],Im(phi_x[i])],i=1..N_x)],color=blue): � |
�
| > | �plots[display](PL1,PL2,view=[-20..10,-0.6..0.6],style=point); � |
�
![[Maple Plot]](images/WignerDiscrete102.gif) �
�
�
| > | �dx*add(abs(phi_x[i])^2,i=1..N_x); � |
�
![]() �
�
� The wavepacket is normalized. �
�� Now the problem of carrying out the Wigner transform becomes one of bookkeeping: we pick � x � -values on the mesh, and read ranges of � s � -values into the appropriate arrays for the FT. Let us pick some � x � -values first. �
��
| > | �Np:=20: i_x_arr:=[seq(210+i*2,i=1..Np)]; � |
�
![]() �
�
�
| > | �seq(xv[i_x_arr[i]],i=1..nops(i_x_arr)); � |
�
![]() �
�
� Let's do a single � x � -value: �
��
| > | �xval:=xv[230]; � |
�
![]() �
�
� A technical point: while we assemble the array for the Fourier transform we have to keep in mind how the � s � -array is filled. In the previous application of the FFT algorithm the � s � -values were substituted. Here we just want to use look-up from the � phi_x � array. The next line serves as a reminder: �
��
| > | �#sv[i]:=evalhf((i-1)*ds): sv[N-i+1]:=evalhf(-i*ds): � |
�
| > | �yR:=array(1..N): yI:=array(1..N): � |
�
| > | �i_x:=230; for i from 1 to N/2 do: rho:=evalc(phi_x[i_x+(i-1)]*conjugate(phi_x[i_x-(i-1)])); yR[i]:=evalf(Re(rho)); yI[i]:=evalf(Im(rho)); rho:=evalc(phi_x[i_x-i]*conjugate(phi_x[i_x+i])); yR[(N-i+1)]:=evalf(Re(rho)); yI[(N-i+1)]:=evalf(Im(rho)); od: � |
�
![]() �
�
�
| > | �plot([seq([sv[i],yR[i]],i=1..N)],style=point); � |
�
![[Maple Plot]](images/WignerDiscrete108.gif) �
�
�
| > | �plot([seq([sv[i],yI[i]],i=1..N)],style=point,color=blue); � |
�
![[Maple Plot]](images/WignerDiscrete109.gif) �
�
�
| > | �N_s:=ds*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
![]() �
�
�
| > | �evalhf(FFT(n, var(yR), var(yI))); � |
�
![]() �
�
�
| > | �N_p:=dp*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
![]() �
�
� The imaginary part should vanish; is this true? �
��
| > | �dp*add(yI[i]^2,i=1..N); � |
�
![]() �
�
�
| > | �yRn:=array(1..N): sq:=evalf(1/sqrt(2*Pi)): for i from 1 to N do: yRn[i]:=ds*sq*yR[i]: od: � |
�
| > | �N_p:=dp*add(yRn[i]^2,i=1..N); � |
�
![]() �
�
�
| > | �plot([seq([pv[i],yRn[i]],i=1..N)],style=line); � |
�
![[Maple Plot]](images/WignerDiscrete115.gif) �
�
� Now that we know how it works, we can put it into a loop, and try a time-evolved wavefunction: �
��
| > | �phi:=No*subs(t=10,psi): � |
�
| > | �N_x:=512: dx:=0.4: xv:=array(1..N_x): phi_x:=array(1..N_x): for i from 1 to N_x do: xv[i]:=(i-N_x/2)*dx: phi_x[i]:=evalf(subs(x=xv[i],phi)): od: � |
�
| > | �PlR:=plot([seq([xv[i],Re(phi_x[i])],i=1..N_x)],color=red,style=point): � |
�
| > | �PlI:=plot([seq([xv[i],Im(phi_x[i])],i=1..N_x)],color=blue,style=point): � |
�
| > | �plots[display]({PlR,PlI},view=[-15..20,-0.5..0.5]); � |
�
![[Maple Plot]](images/WignerDiscrete116.gif) �
�
� We adjust the computational mesh of � x � -values at which the Wigner function � f(x,p) � is calculated to span interesting regions in � x � -space. �
��
| > | �Np:=35: i_x_arr:=[seq(220+i*2,i=1..Np)]; � |
�
![]() �
�
![]() �
�
�
| > | �seq(xv[i_x_arr[i]],i=1..nops(i_x_arr)); � |
�
![]() �
�
![]() �
�
� In the loop over the � x � -mesh we verify in the � Tnorm � variable that the overall norm of the Wigner distribution approximately equals unity. �
��
| > | �Tnorm:=0: for k from 1 to Np do: i_x:=i_x_arr[k]: xval:=xv[i_x]; � |
�
| > | �yR:=array(1..N): yI:=array(1..N): � |
�
| > | �for i from 1 to N/2 do: rho:=evalc(phi_x[i_x+(i-1)]*conjugate(phi_x[i_x-(i-1)])); yR[i]:=evalf(Re(rho)); yI[i]:=evalf(Im(rho)); rho:=evalc(phi_x[i_x-i]*conjugate(phi_x[i_x+i])); yR[(N-i+1)]:=evalf(Re(rho)); yI[(N-i+1)]:=evalf(Im(rho)); od: � |
�
| > | �N_s:=ds*add(yR[i]^2+yI[i]^2,i=1..N); � |
�
| > | �evalhf(FFT(n, var(yR), var(yI))); � |
�
| > | �yRn:=array(1..N): sq:=evalf(1/sqrt(2*Pi)): for i from 1 to N do: yRn[i]:=ds*sq*yR[i]: od: � |
�
| > | �N_p:=dp*add(yRn[i]^2+ds*sq*yI[i]^2,i=1..N); print("xval,Ns,Np=",xval,N_s,N_p); Tnorm:=Tnorm+(xv[i_x_arr[2]]-xv[i_x_arr[1]])*N_p; � |
�
| > | �PL[k]:=plot([seq([pv[i],yRn[i]],i=1..N)],style=line,title=cat("x= ",trunc(10*xval),"/10")): od: Tnorm; � |
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
![]() �
�
�
| > | �plots[display]([seq(PL[i],i=1..Np)],insequence=true); � |
�
![[Maple Plot]](images/WignerDiscrete157.gif) �
�
�
| > | �plots[display](PL[20]); � |
�
�
�
![[Maple Plot]](images/WignerDiscrete158.gif) �
�
� Exercise 2: �
�� Explore the Wigner distribution using the cuts as shown above for different times, and describe the behaviour. Negative parts in the distribution are apparently strong when the backscattered and forward-moving parts of the wavepacket meet and interfere, but also apparently behind the barrier, where no interference takes place. �
��
�� Exercise 3: �
�� Vary the wavepacket parameters (and the potential parameters), and explore the Wigner distribution. For wavepackets moving rapidly over the barrier the regions of negative Wigner functions should be small. What happens for scattering from attractive wells? �
��
�� The Maple program can also be used to read in wavepackets produced with Fortran or C programs which employ an FFT-based time integrator (such as the one illustrated in � Ehrenfest.mws � ). To explore Wigner distributions for such situations can make up an interesting student project. �
��
| > | �