Research in The Audette Lab
Research in our group focuses on understanding how microorganisms utilize sophisticated purpose-built nanomachines for transferring genetic material and effector molecules across membranes, and facilitate adherence to a variety of surfaces. Understanding of how these nanosystems are assembled from their component proteins, as well as their effects on infection and the development of resistance strategies, is critical to the development of more streamlined approaches to dealing with infection and drug resistance in pathogenic organisms. In addition, understanding how these systems function at a structural level is central to the development of biological nanosystems for applications as biosensors.
NEW - Some of our recent research into the pilin-derived protein nanotubes has been featured in BioMed Central's online magazine Biome.
Structural and Functional Studies of Proteins Involved in Bacterial Conjugation
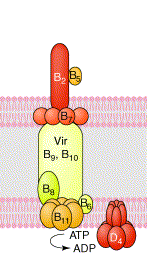
A representation of the T4SS from A. tumefaciens. Adapted from Remaut
& Waksman, Curr. Opin. Struct. Biol. 14: 161-170 (2004).
We have targeted several proteins unique to the F-plasmid system, as well as several which have homologs in other systems, and are looking primarily at protein structure, function and protein-protein interactions. These studies are aimed at understanding not only the structures and functions of the components that form the conjugation apparatus, but also the ways these proteins interact to form a complex molecular pump to transfer DNA from the donor to the recipient cell.
Bacterial conjugation is the process by which DNA can be transferred from one bacterium (donor) to another (recipient). This process enables the distribution of genetic material throughout a bacterial population, and is a mechanism for bacterial evolution and adaptaion to unqiue environments. It is also a mechanisms by which genes encoding antibiotic resistance and other virulence factors amongst are spread through a bacterial population. Related systems are also used by several pathogens to secrete virulence factors into the medium (for instance the toxin from B. pertussis) or to directly inject them into host cells (for instance CagA from the human pathogen H. pylori or the T-DNA of the plant pathogen A. tumefaciens). These systems have been classified a subgroup of the type IV secretion system (T4SS) based upon the similarities between the genes involved in the processes.
The F-plasmid of E. coli, first described in 1946, is the prototypical system for bacterial conjugation. However, despite a large accumulation of genetic and biochemical data over the last half century, a detailed structural analysis of the component proteins of this system remains elusive. Our research is focused on examining the component proteins that make up the F-plasmid T4SS from a structural and functional standpoint. We are employing several biochemical and biophysical methodologies in our studies, with protein X-ray crystallography being our primary research tool.
Protein-based Nanotechnology: Pilin-Derived Protein Nanotubes
We have shown that a highly soluble engineered pilins from P. aeruginosa can oligomerize in solution (Audette et al. Nano Lett. 4(10), 1897-1902 (2004)) and at surfaces (Lombardo et al. J. Bionanosci. 3(1), 61-65 (2009)). The structures derived from these engineered pilins are in effect protein nanotubes (PNTs), and are interesting targets for bionanotechnolgy applications such as biosensors.
We are currenlty focusing on understanding the mechanisms of how PNTs oligomerize from the monomeric protein precursor, and are investigating the development of these novel PNTs through protein engineering. For example, what are the kinetic parameters of PNT formation? Can we rationally modify the monomeric pilin protein to modify the assembly and kinetics of PNT formation? Also, can we add specific functionality to either the inside or outside of the PNT through modification of the monomeric protein, or by using them as an adapter technology for a biologically relevant nanomachine?
We are also characetrizing the assembly systems and T4P/PNTs from a variety of bacteria, with an aim at a greater understanding of how these structures are assembled, the specific interactions that occur at the binding interface during surface adherence, and the development of pilin-derived PNTs for applications in biosensors and bionanoelectronics.

Pilin-derived protien nanotubes (PNTs) oligomerized from self-assembled alkylthiol monolayers on an Au(111) surface.
From: Lombardo et al. J. Bionanosci., 3(1), 61-65 (2009).
Another focus of our research program is the type II secretion system (T2SS) that assembles the type IV pilus (T4P) in several organisms, including Pseudomonas aeruginosa. Bacteria such as P. aeruginosa use T4P, which are fibrous assemblies of pilin monomers, during the initial stages of infection to attach to host cells.
Finally, I am interested in the development of methods which allow crystallographers to predict potential amino acid mutations in order to improve the overall quality of protein crystals. The ability to obtain high quality crystals for X-ray diffraction is a significant hurdle for many researchers. The goal of this research is to develop a rational approach to identify potential mutations for a researchers protein of interest using moderate resolution X-ray diffraction data in order to obtain higher resolutions, and allow for a more confident analysis of crystallographic results.
We also use linux as our general operating system on our computers (with some exceptions). There are a variety of reasons for this, including security, data handling, crystallographic software developments etc.. Follow this link to get an idea of some linux basics.
We are also interested in the correlation of multiple biophysical techniques in the examination of protein structure and function. For example, in addition to X-ray crystallography, biophysical techniques such as Nuclear Magnetic Resonance (NMR) spectroscopy, Small Angle X-ray Scattering (SAXS), Multi-Angle Light Scattering (MALS), Electron Microscopy (EM) and fibre diffraction can provide significant insights into protein structure and dynamics and can complement well designed functional studies. Each distinct methodology has its own respective strengths, and the coupling of these methods can only benefit our understanding into the relation between protein structure and biochemical function. Wherever possible, all studies in our lab employ a variety of biophysical methods, while primarily employing X-ray crystallography, in order to address our research goals.